Tutorial
In the "Search"page, you can use the "Submit" button on the "Search" page to find specific variants by Uniprot ID, gene name or protein name. You can also click example links to perform an example search.

Examples:
P11487- Search by UniProt IDFGF3- Search by gene nameCyclin-O- Search by protein name
In the "Search results" page, you can click the "Download" button to get the data in txt formats. Explanation of the columns are shown by clicking the icon. The detailed information about a specific variant are displayed by clicking any "Mutation site", "P. Level", "Wild type -> Mutation" or the icon.
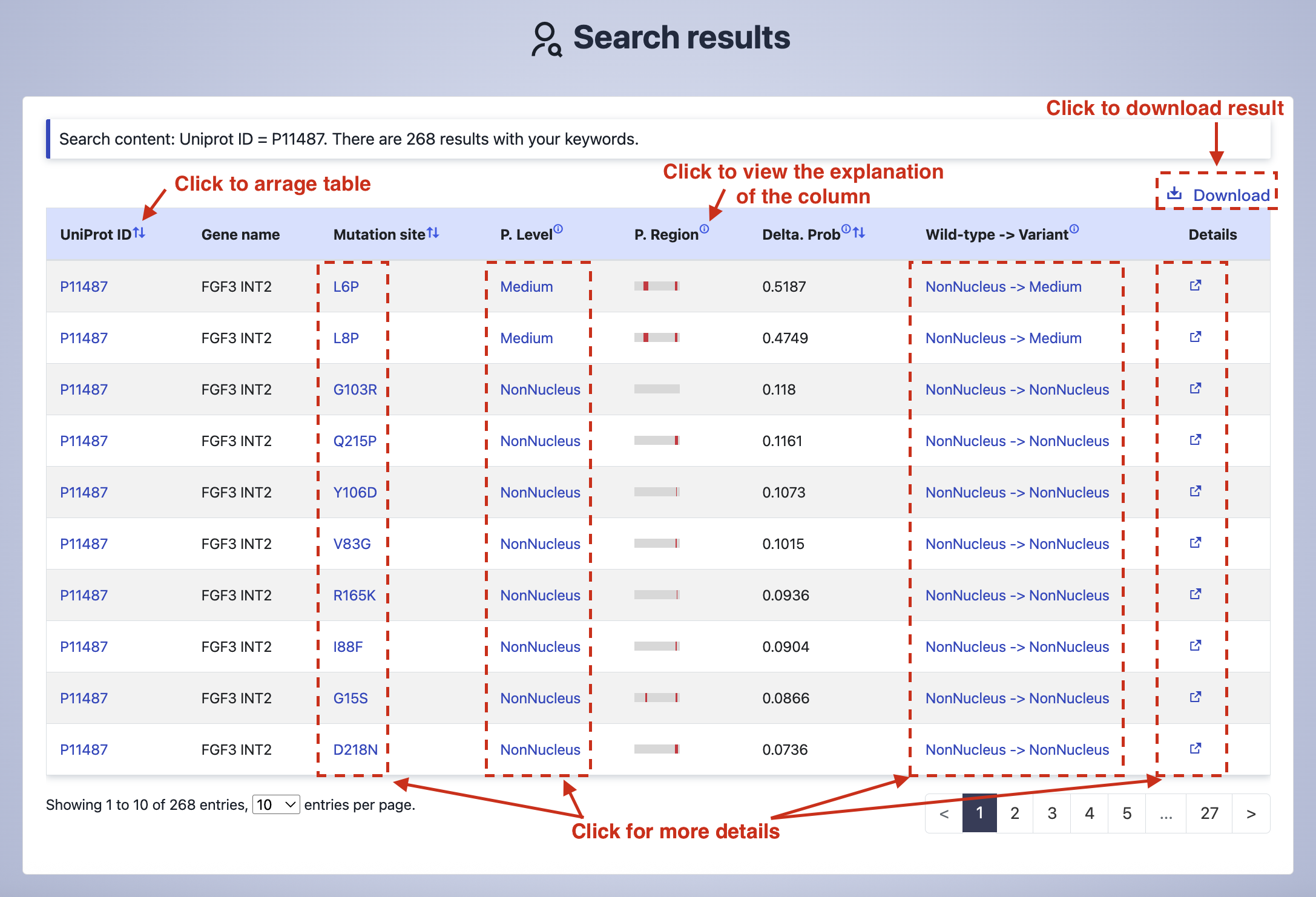
Column explanation:
P. Level- The nuclear localization level predicted by pSAM, categorized as High (≥0.737), Medium (0.476-0.736), Low (0.328-0.475), and NonNucleus (<0.328) based on nuclear localization probability thresholds.P. Region- The determinants for nuclear localization (DNL) of mutant proteins predicted by pSAM. The red region represents the DNL region.Delta. Prob- The difference value between the mutant and wild type proteins in the nuclear localization probabilities predicted by pSAM.Wild type -> Mutation- The difference in nuclear localization probability predicted by pSAM between mutant and wild-type proteins, calculated as mutant probability minus wild-type probability.
The "Detailed information" page consists of five parts: "About Protein", "Variation", "Nuclear Localization Codes", "Subcellular Location" and "Structure".
(1) About Protein
The basic information of the protein.

(2) Variation
The disease and functional annotation of the mutation site.
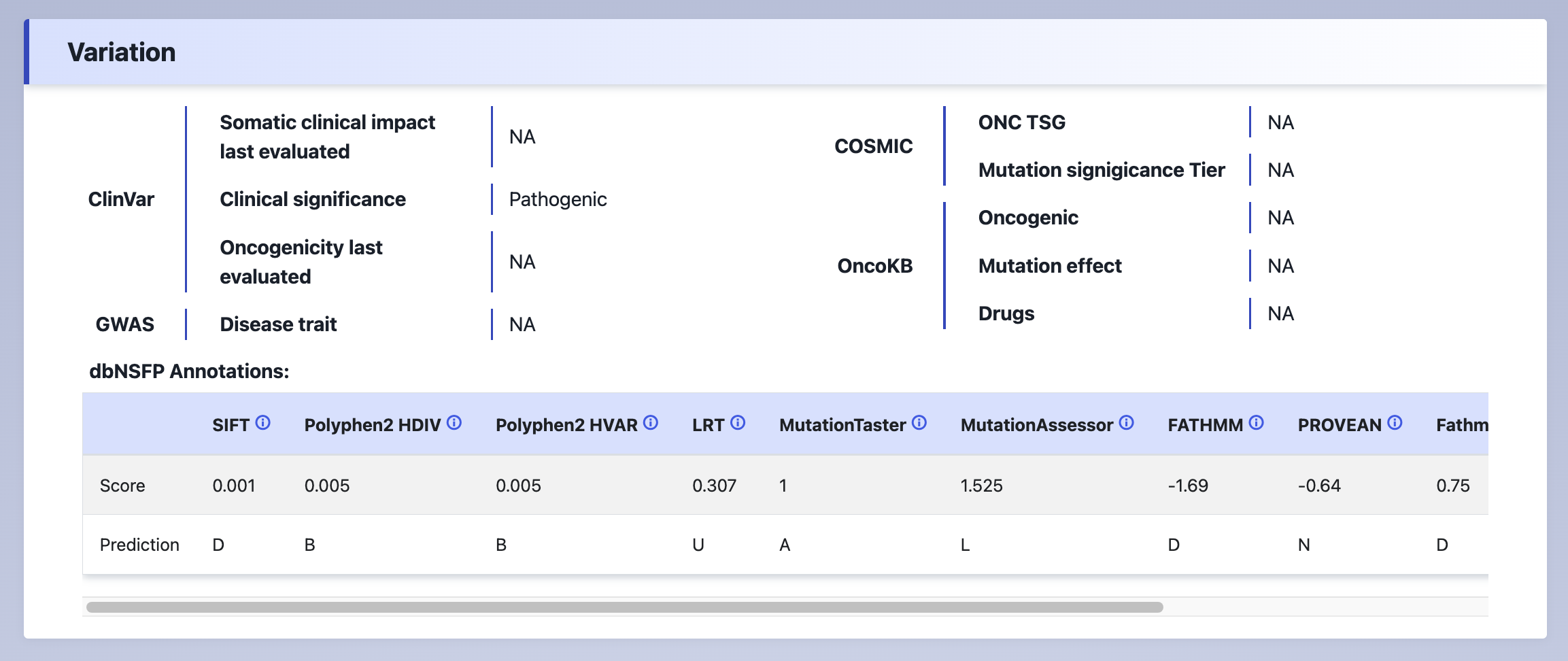
Explanation:
Somatic clinical impact last evaluated- The most recent date when the somatic clinical impact was evaluated.Clinical significance- The clinical significance of the mutation. Asssertion criteria includes: pathogenic, likely pathogenic, uncertain significance, benign, likely benign.Oncogenicity last evaluated- The most recent date of oncogenicity evaluation.ONC TSG- Classification of COSMIC as oncogene or tumor suppressor gene, depending on its somatic mutation profile and functional role in oncogenesis.Mutation signigicance Tier- The tier of mutation significance based on the COSMIC database.Oncogenic- Oncogenic activity classification.Mutation effect- Biological consequences of mutations annotated.Drugs- Targeted drugs/therapies curated by OncoKB.dbNSFP annotation- 11 tools for predicting the effect of mutations on function from dbNSFP database.
(3) Nuclear Localization Codes
The nuclear localization codes predicted by pSAM and NLS/NES annotation collected from various sources.
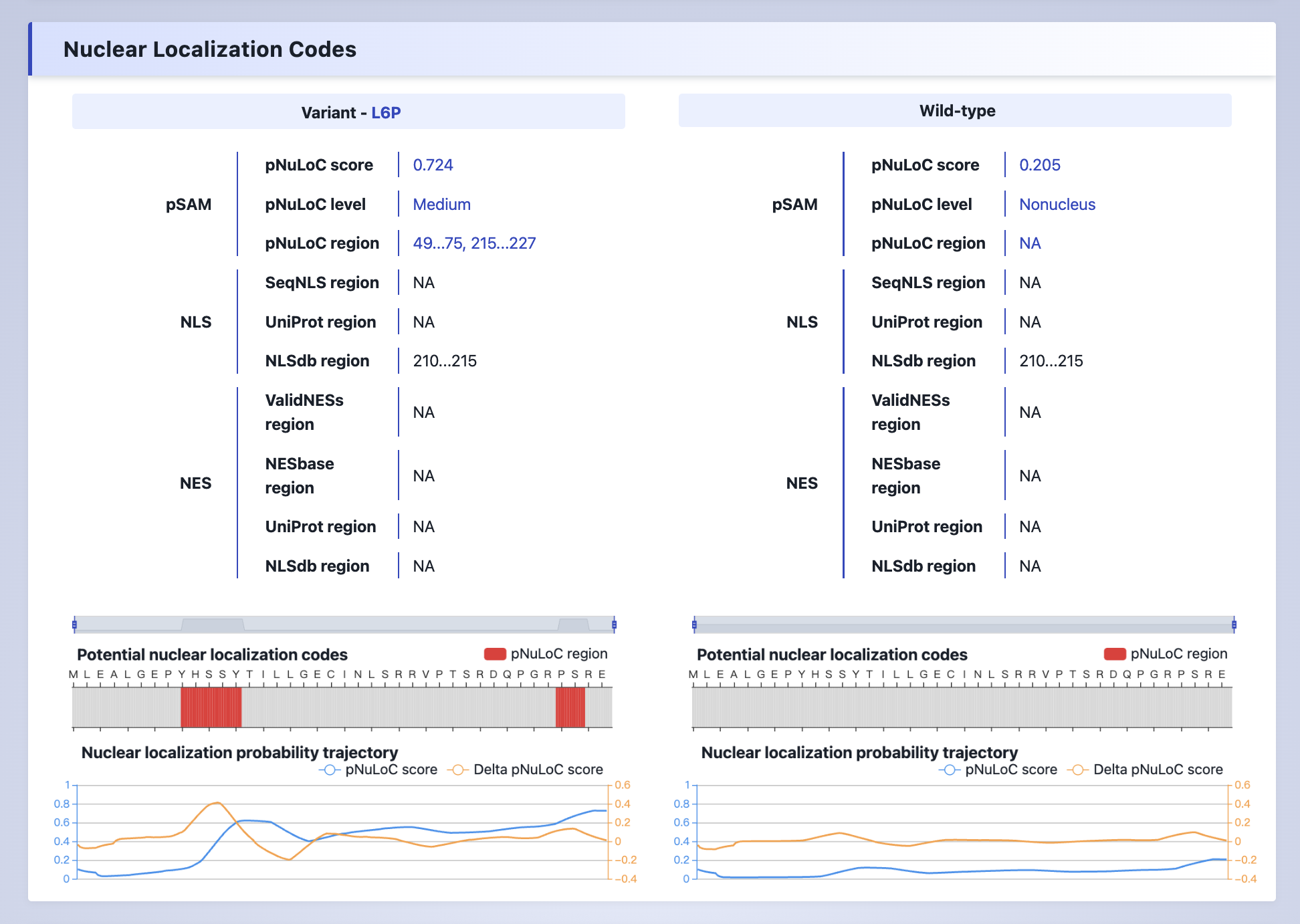
Explanation:
pNuLoC score- The nuclear localization probability predicted by pSAM.pNuLoC level- The nuclear localization level predicted by pSAM, categorized as High (≥0.737), Medium (0.476-0.736), Low (0.328-0.475), and NonNucleus (<0.328) based on nuclear localization probability thresholds.pNuLoC Region- The determinants for nuclear localization (DNL) predicted by pSAM.SeqNLS region- The experimental validataed NLS region collected from SeqNLS database.UniProt region- The experimental validataed NLS or NES region collected from UniProt database.NLSbase region- The predicted NLS or NES region collected from NLSbase database.ValidNESs region- The experimental validataed NES region collected from ValidNESs database.NESbase region- The experimental validataed NES region collected from NESbase database.Poteintial nuclear localization codes- The determiants for nuclear localization predicted by pSAM.Nuclear localization probability trajectory- The nuclear localization probability trajectory predicted by pSAM, see the paper for details.
(4) Subcellular Location
The subcellular localization information of the protein.
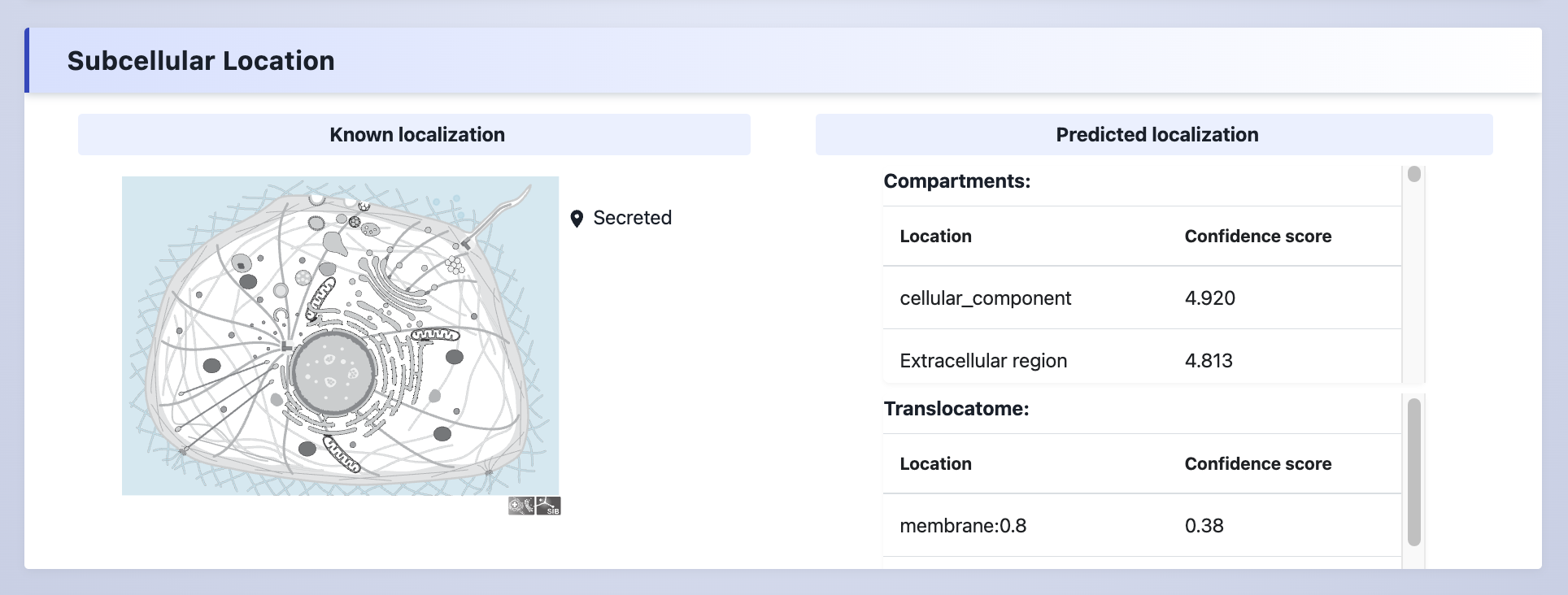
Explanation:
Known subcellular location- The known subcellular location data was collected from UniProt database.Predicted subcellular location- Compartments predicted subcellular locations for proteins. Translocatome predicted translocating proteins.
(5) Structure
The PTM information, disorder values, surface accessibility, polar, charge and hydropathy information, secondary structure, and 3D-structure of the protein.
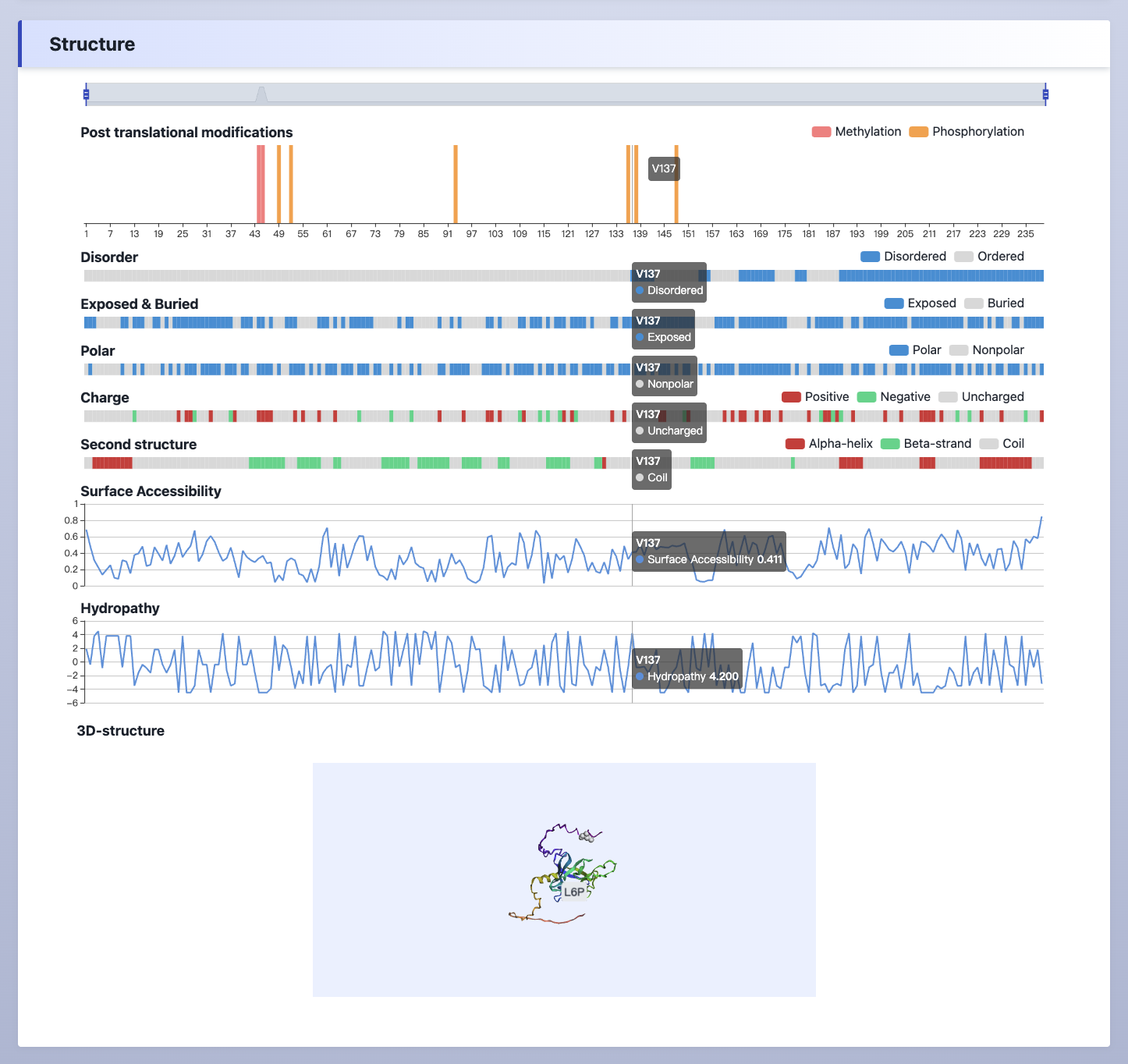
Explanation:
Post translational modifications- The post translational modifications information was collected from qPTM database.Disorder, Exposed & Buried- The disorder values were calculated by IUPred.Surface accessibility, Second structure- The surface accessibility values were predicted by NetSurfP.Polar, Charge, Hydropathy- The polar, charge and hydropathy information were retrived from AAindex.3D-structure- The 3D-structure of the protein retrived from PDB database.
NuLocVar data will be available after publication.
